当前位置:
X-MOL 学术
›
Water Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Identification of the microorganisms for methane-dependent arsenate reduction in wetland using DNA-stable isotope probing and metagenomics
Water Research ( IF 11.4 ) Pub Date : 2025-05-30 , DOI: 10.1016/j.watres.2025.123934
Xiaoxiao Yu, Jibing Li, Yujie Zhou, Yun Chen, Lina Zou, Chunling Luo, Chaofeng Shen, Fengjie Liu, Jianming Xu, Xianjin Tang
Water Research ( IF 11.4 ) Pub Date : 2025-05-30 , DOI: 10.1016/j.watres.2025.123934
Xiaoxiao Yu, Jibing Li, Yujie Zhou, Yun Chen, Lina Zou, Chunling Luo, Chaofeng Shen, Fengjie Liu, Jianming Xu, Xianjin Tang
|
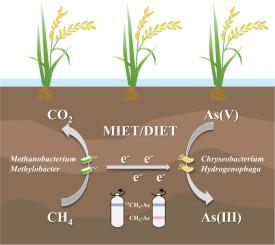
The methane-dependent arsenate reduction (M-AsR) process is recently demonstrated to enhance the release of mobile and toxic arsenite [As(III)], with critical implications for ecosystem safety in wetland ecosystem. However, the key functional microorganisms and the underlying metabolic mechanisms of M-AsR in wetland remain unclear. In this study, 13C-labeled methane (CH4) was used as the sole carbon source to track the active microorganisms responsible for M-AsR. DNA-stable isotope probing (DNA-SIP) combined with amplicon and metagenomic sequencing was further employed to identify the microorganisms involved in M-AsR. The results showed that arsenate [As(V)] reduction occurred exclusively in the treatment amended with both CH₄ and As(V). After a 50-day incubation, significant shifts in the relative abundance of functional genes (pmoA, ANME-2d mcrA and arrA) were observed in the heavy DNA fractions from the treatment amended with 13CH4 and As(V), indicating the incorporation of 13C into M-AsR microorganisms. Furthermore, the Methanobacterium, Methylobacter and arsenate-reducing bacteria (Chryseobacterium and Hydrogenophaga) were the predominant genera in 13CH4-As heavy fractions and identified as the potential microorganisms responsible for M-AsR in wetland. Metagenomic analysis further confirmed that most of these microorganisms contained genes for CH4 oxidation and As(V) reduction. This multi-omics approach provides mechanistic insights into microbial mediated As(V) reduction in methane-rich wetland area.
中文翻译:

使用 DNA 稳定同位素探测和宏基因组学鉴定湿地中甲烷依赖性砷酸盐减少的微生物
最近证明,甲烷依赖性砷酸盐还原 (M-AsR) 过程可以增强移动和有毒亚砷酸盐 [As(III)] 的释放,对湿地生态系统的生态系统安全具有重要意义。然而,湿地中 M-AsR 的关键功能微生物及其潜在的代谢机制仍不清楚。在这项研究中,13 个 C 标记的甲烷 (CH4) 被用作唯一的碳源来追踪负责 M-AsR 的活性微生物。进一步采用 DNA 稳定同位素探测 (DNA-SIP) 结合扩增子和宏基因组测序来鉴定参与 M-AsR 的微生物。结果表明,砷酸盐 [As(V)] 的减少仅发生在用 CH₄ 和 As(V) 修正的处理中。孵育 50 天后,在用 13CH4 和 As(V) 修饰的处理的重 DNA 组分中观察到功能基因 (pmoA、ANME-2d mcrA 和 arrA) 的相对丰度发生显着变化,表明 13C 掺入 M-AsR 微生物中。此外, 甲烷杆菌属、甲基杆菌属和砷酸盐还原菌 (Chryseobacterium 和 Hydrogenophaga) 是 13CH4-As 重组分中的优势属,并被确定为导致湿地 M-AsR 的潜在微生物。宏基因组分析进一步证实,这些微生物中的大多数都含有 CH4 氧化和 As(V) 还原的基因。这种多组学方法为微生物介导的富含甲烷的湿地地区 As(V) 减少提供了机制见解。
更新日期:2025-05-31
中文翻译:

使用 DNA 稳定同位素探测和宏基因组学鉴定湿地中甲烷依赖性砷酸盐减少的微生物
最近证明,甲烷依赖性砷酸盐还原 (M-AsR) 过程可以增强移动和有毒亚砷酸盐 [As(III)] 的释放,对湿地生态系统的生态系统安全具有重要意义。然而,湿地中 M-AsR 的关键功能微生物及其潜在的代谢机制仍不清楚。在这项研究中,13 个 C 标记的甲烷 (CH4) 被用作唯一的碳源来追踪负责 M-AsR 的活性微生物。进一步采用 DNA 稳定同位素探测 (DNA-SIP) 结合扩增子和宏基因组测序来鉴定参与 M-AsR 的微生物。结果表明,砷酸盐 [As(V)] 的减少仅发生在用 CH₄ 和 As(V) 修正的处理中。孵育 50 天后,在用 13CH4 和 As(V) 修饰的处理的重 DNA 组分中观察到功能基因 (pmoA、ANME-2d mcrA 和 arrA) 的相对丰度发生显着变化,表明 13C 掺入 M-AsR 微生物中。此外, 甲烷杆菌属、甲基杆菌属和砷酸盐还原菌 (Chryseobacterium 和 Hydrogenophaga) 是 13CH4-As 重组分中的优势属,并被确定为导致湿地 M-AsR 的潜在微生物。宏基因组分析进一步证实,这些微生物中的大多数都含有 CH4 氧化和 As(V) 还原的基因。这种多组学方法为微生物介导的富含甲烷的湿地地区 As(V) 减少提供了机制见解。


















































 京公网安备 11010802027423号
京公网安备 11010802027423号