当前位置:
X-MOL 学术
›
Water Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Combining analysis of individual and wastewater whole genome sequencing improves SARS-CoV-2 surveillance
Water Research ( IF 11.4 ) Pub Date : 2025-06-03 , DOI: 10.1016/j.watres.2025.123953
Evan P. Troendle, Andrew J. Lee, Marina I. Reyne, Danielle M. Allen, Stephen J. Bridgett, Clara H. Radulescu, Michael Glenn, John-Paul Wilkins, Francesco Rubino, Behnam Firoozi Nejad, Cormac McSparron, Marc Niebel, Derek J. Fairley, Kate E. Binley, Christopher J. Creevey, Jennifer M. McKinley, Timofey Skvortsov, Deirdre F. Gilpin, John W. McGrath, Connor G.G. Bamford, David A. Simpson
Water Research ( IF 11.4 ) Pub Date : 2025-06-03 , DOI: 10.1016/j.watres.2025.123953
Evan P. Troendle, Andrew J. Lee, Marina I. Reyne, Danielle M. Allen, Stephen J. Bridgett, Clara H. Radulescu, Michael Glenn, John-Paul Wilkins, Francesco Rubino, Behnam Firoozi Nejad, Cormac McSparron, Marc Niebel, Derek J. Fairley, Kate E. Binley, Christopher J. Creevey, Jennifer M. McKinley, Timofey Skvortsov, Deirdre F. Gilpin, John W. McGrath, Connor G.G. Bamford, David A. Simpson
|
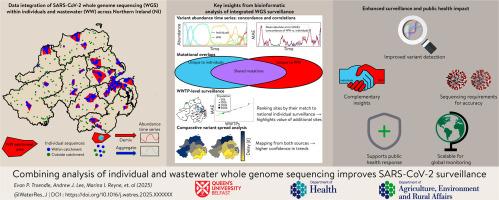
Effective pathogen surveillance is critical for public health decision-making, with both individual and environmental monitoring playing essential roles. While wastewater (WW) and individual whole genome sequencing (WGS) have been used to monitor SARS-CoV-2 dynamics, their complementary potential for enhancing national-level genomic surveillance remains underexplored. This study aimed to evaluate the unique and combined contributions of WW and individual WGS to genomic surveillance. We conducted SARS-CoV-2 WGS on over 4,000 WW samples and 23,000 individual samples across Northern Ireland (NI) between 2021 and 2023. SARS-CoV-2 RNA was amplified using the ARTIC nCov-2019 and Mini-XT protocols and sequenced on Illumina MiSeq. Variant compositions in WW data were analysed using Freyja and compared to individual data using time series analysis, correlation assessments, and volatility measurements via numerical derivatives, with mean absolute error (MAE) calculations used to assess concordance. Wastewater treatment plants (WWTPs) were ranked by concordance to individual WGS data. WW and individual WGS complementarity was quantified by mutation classification and overlap analysis. Temporal curve shifting was used to identify lags or leads in variant detection and to infer differences in geospatial spread between WW and individual sequencing data.We confirmed strong concordance between WW and individual variant compositions (mean MAE = 6.2%). MAE was inversely correlated with sequencing rate (Pearson r=−0.37, p<0.001) and increased during periods with more circulating variants, highlighting the value of increased sequencing efforts during volatile periods. The population size served by a WWTP was not a reliable indicator of how well its variant composition matched that of the national individual sequencing programme. Both individual and WW-based sequencing (WBS) detected unique, as well as common mutations. Patterns of variant spread within NI were consistent between both programmes (Pearson r=0.63, p = 0.036), providing complementary insights into variant trends and geospatial spread.We demonstrate that integration of individual and WW WGS data offers more comprehensive SARS-CoV-2 genomic surveillance and improves confidence in predictions of variant composition and spread.
中文翻译:

将个体测序和废水全基因组测序分析相结合,可改善 SARS-CoV-2 监测
有效的病原体监测对于公共卫生决策至关重要,个人和环境监测都发挥着至关重要的作用。虽然废水 (WW) 和个体全基因组测序 (WGS) 已被用于监测 SARS-CoV-2 动态,但它们在加强国家级基因组监测方面的互补潜力仍未得到充分开发。本研究旨在评估 WW 和个体 WGS 对基因组监测的独特和综合贡献。2021 年至 2023 年期间,我们对北爱尔兰 (NI) 的 4,000 多个 WW 样本和 23,000 个个体样本进行了 SARS-CoV-2 WGS。使用 ARTIC nCov-2019 和 Mini-XT 方案扩增 SARS-CoV-2 RNA,并在 Illumina MiSeq 上测序。使用 Freyja 分析 WW 数据中的变体组成,并使用时间序列分析、相关性评估和通过数值导数的波动性测量与单个数据进行比较,并使用平均绝对误差 (MAE) 计算来评估一致性。废水处理厂 (WWTP) 根据与单个 WGS 数据的一致性进行排名。通过突变分类和重叠分析量化 WW 和个体 WGS 互补性。时间曲线偏移用于识别变异检测的滞后或领先优势,并推断 WW 和单个测序数据之间地理空间传播的差异。我们证实 WW 和个体变体组成之间具有很强的一致性 (平均 MAE = 6.2%)。MAE 与测序率呈负相关 (Pearson r=-0.37,p<0.001),并且在流通变异较多的时期增加,突出了在波动时期增加测序工作的价值。 污水处理厂所服务的种群规模并不是其变体组成与国家个体测序计划的匹配程度的可靠指标。个体测序和基于 WW 的测序 (WBS) 均检测到独特突变和常见突变。两个项目之间 NI 内部的变体传播模式是一致的 (Pearson r=0.63,p = 0.036),为变体趋势和地理空间传播提供了互补的见解。我们证明,个体和 WW WGS 数据的整合提供了更全面的 SARS-CoV-2 基因组监测,并提高了预测变体组成和传播的可信度。
更新日期:2025-06-04
中文翻译:

将个体测序和废水全基因组测序分析相结合,可改善 SARS-CoV-2 监测
有效的病原体监测对于公共卫生决策至关重要,个人和环境监测都发挥着至关重要的作用。虽然废水 (WW) 和个体全基因组测序 (WGS) 已被用于监测 SARS-CoV-2 动态,但它们在加强国家级基因组监测方面的互补潜力仍未得到充分开发。本研究旨在评估 WW 和个体 WGS 对基因组监测的独特和综合贡献。2021 年至 2023 年期间,我们对北爱尔兰 (NI) 的 4,000 多个 WW 样本和 23,000 个个体样本进行了 SARS-CoV-2 WGS。使用 ARTIC nCov-2019 和 Mini-XT 方案扩增 SARS-CoV-2 RNA,并在 Illumina MiSeq 上测序。使用 Freyja 分析 WW 数据中的变体组成,并使用时间序列分析、相关性评估和通过数值导数的波动性测量与单个数据进行比较,并使用平均绝对误差 (MAE) 计算来评估一致性。废水处理厂 (WWTP) 根据与单个 WGS 数据的一致性进行排名。通过突变分类和重叠分析量化 WW 和个体 WGS 互补性。时间曲线偏移用于识别变异检测的滞后或领先优势,并推断 WW 和单个测序数据之间地理空间传播的差异。我们证实 WW 和个体变体组成之间具有很强的一致性 (平均 MAE = 6.2%)。MAE 与测序率呈负相关 (Pearson r=-0.37,p<0.001),并且在流通变异较多的时期增加,突出了在波动时期增加测序工作的价值。 污水处理厂所服务的种群规模并不是其变体组成与国家个体测序计划的匹配程度的可靠指标。个体测序和基于 WW 的测序 (WBS) 均检测到独特突变和常见突变。两个项目之间 NI 内部的变体传播模式是一致的 (Pearson r=0.63,p = 0.036),为变体趋势和地理空间传播提供了互补的见解。我们证明,个体和 WW WGS 数据的整合提供了更全面的 SARS-CoV-2 基因组监测,并提高了预测变体组成和传播的可信度。


















































 京公网安备 11010802027423号
京公网安备 11010802027423号